|
|
|
|
|
胡家志研究组开发出优化基因编辑和追踪DNA修复的新方法 | Cell Discovery |
|
|
论文标题:Optimizing genome editing strategy by primer-extension-mediated sequencing
期刊:Cell Discovery
作者:Jianhang Yin, Mengzhu Liu, Yang Liu, Jinchun Wu, Tingting Gan, Weiwei Zhang, Yinghui Li, Yaxuan Zhou, Jiazhi Hu
发表时间:2019/03/26
数字识别码: 10.1038/s41421-019-0088-8
原文链接:http://t.cn/EoQdjQm
微信链接:https://mp.weixin.qq.com/s/kYkJstMCuftv7S2ddQ_eJQ
2019年3月26日,北京大学
学院和北大-清华
联合中心胡家志课题组在Cell Discovery 上发表题为Optimizing genome editing strategy by primer-extension-mediated sequencing的论文。该研究描述了一种新的方法,可以用来同时定量检测Cas9编辑效率和脱靶活性以及编辑引起的染色体异常结构,即primer-extension-mediated sequencing(PEM-seq)。这是对基因编辑和DNA损伤修复等领域都有巨大促进作用的新技术。
CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats and CRISPR-associated proteins)是目前最常用的基因编辑工具酶,在科研领域被广泛应用,而在临床方面的应用因为其脱靶活性一度停滞不前。它通过guide RNA(gRNA)与靶向DNA序列的配对,从而将Cas9锚定在靶向基因并诱导产生DNA双链断裂(DSB)。在基因编辑诱发的DSB的修复过程中,一定几率会产生基因突变或者外源DNA片段的插入,从而达到基因编辑的目的。在实际应用过程中,一个好的基因编辑酶Cas9需要同时满足高效切割靶位点、低脱靶活性和低染色体异常三个特点。目前在这三个方面,有一部分基于PCR的方法用于估算基因编辑效率,但其结果可靠性有待提高;有一些基于高通量测序的方法可以在体内或者体外检测基因编辑酶的脱靶活性;尚无系统的可定量的测量基因组异常结构的方法。
依据DNA双链断裂与染色体易位的原理,胡家志课题组在已有的高通量测序方法(Hu et al., Nature Protocols 2016)的基础上开发了灵敏度更高且可以全面且定量评估基因编辑的新方法 PEM-seq。与以往基于二代测序评估Cas9脱靶活性的方法相比,PEM-seq不仅可以灵敏的找出Cas9的脱靶位点,还可以精确定量CRISPR/Cas9在靶向位点的切割效率,从而找到更加高效安全的Cas9切割位点。与此同时,PEM-seq还深度揭示了靶向位点附近由于基因编辑而产生的染色体异常结构,例如大片段缺失、染色体易位等。以靶向治疗RAG1基因上的RAG1A编辑位点为例,胡家志课题组发现:在Cas9切割位点5kb以内存在着占总编辑事件2.5%的大量的染色体缺失倒位等异常结构;更为严重的是,这些异常结构可以延伸至距离切割位点50kb甚至更长的领域,足以严重威胁基因组的稳定性。这些现象揭示了Cas9应用前评估的必要性。应用PEM-seq可以全面地评估Cas9的切割效率及其所导致的大多异常结构,从而优化基因编辑策略,以达到获得最大编辑效率且尽量减少脱靶活性的效果。
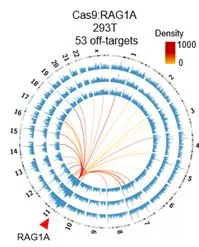
图1: PEM-seq检测出Cas9 在RAG1A位点有53个脱靶位点
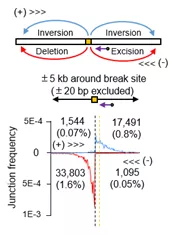
图2:PEM-seq检测到在Cas9切割位点5kb内存在着大量染色体异常机构
同时,为了降低CRISPR/Cas9的脱靶活性,胡家志课题组将现有Cas9变体的突变位点进行了组合筛选,通过PEM-seq筛选出了一个切割效率与野生型Cas9相当但脱靶频率明显更低的变体FeCas9。该酶的使用方法与传统Cas9类似。
此外,PEM-seq在基因组的稳定性研究方面还有较大的潜力。基因组的修复与DNA修复等因素密切相关,而传统的方法多用PCR或者分子克隆获得修复信息,样本小且存在偏好性。PEM-seq与一些之前的高通量测序方法类似,可以提供较大的数据量,且相对而言有一个更加明显的优势,即定量。PEM-seq可以对DNA修复的步骤进行逐步定量,从而细致地描画DNA从损伤到修复的过程,以获得更加精准的模型。
摘要:Efficient and precise genome editing is essential for clinical applications and generating animal models, which requires engineered nucleases with high editing ability while low off-target activity. Here we present a high-throughput sequencing method, primer-extension-mediated sequencing (PEM-seq), to comprehensively assess both editing ability and specificity of engineered nucleases. We showed CRISPR/Cas9-generated breaks could lead to chromosomal translocations and large deletions by PEM-seq. We also found that Cas9 nickase possessed lower off-target activity while with some loss of target cleavage ability. However, high-fidelity Cas9 variants, including both eCas9 and the new FeCas9, could significantly reduce the Cas9 off-target activity with no obvious editing retardation. Moreover, we found AcrIIA4 inhibitor could greatly reduce the activities of Cas9, but off-target loci were not so effectively suppressed as the on-target sites. Therefore, PEM-seq fully evaluating engineered nucleases could help choose better genome editing strategy at given loci than other methods detecting only off-target activity.
阅读论文全文请访问:http://t.cn/EoQdjQm
期刊介绍:Cell Discovery is an open access international journal that publishes results of high significance and broad interest in all areas of molecular and cell biology. Cell Discovery is established in 2015 as a sister journal of the high profile international journal Cell Research
(来源:科学网)
特别声明:本文转载仅仅是出于传播信息的需要,并不意味着代表本网站观点或证实其内容的真实性;如其他媒体、网站或个人从本网站转载使用,须保留本网站注明的“来源”,并自负版权等法律责任;作者如果不希望被转载或者联系转载稿费等事宜,请与我们接洽。