论文标题:Extreme inbreeding in a European ancestry sample from the contemporary UK population
期刊:Nature Communications
作者:Loic Yengo,Naomi R. Wray,Peter M. Visscher
发表时间:2019/09/03
数字识别码:10.1038/s41467-019-11724-6
微信链接:https://mp.weixin.qq.com/s/StETUD2OXZO8lwMk8sD50Q
《自然-通讯》发表的一项研究Extreme inbreeding in a European ancestry sample from the contemporary UK population分析了近亲繁殖及其对健康的潜在影响。
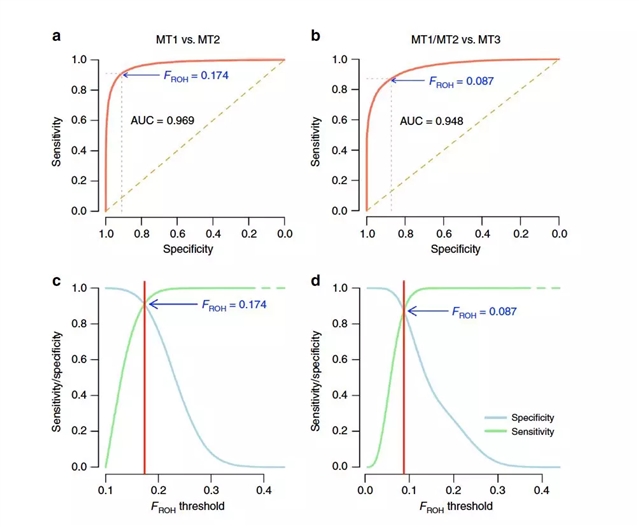
图1 :用来区分不同的近亲繁殖类型的FROH的预测性能 图源:Yengo等
澳大利亚昆士兰大学的Loic Yengo及同事使用英国生物样本库(UK Biobank)中456,414名个体的匿名数据,估算了极端近亲繁殖的普遍性。极端近亲繁殖指一级亲属和二级亲属之间的繁殖,比如父母和子女或其半同胞之间的繁殖。作者以连续性纯合片段(即遗传自父母的相同的基因组片段)为基础展开研究,检测近亲繁殖是否与很多健康后果相关。
作者发现在所有被研究者中,有125人的遗传数据表明他们是一级亲属或二级亲属近亲繁殖的后代。不仅如此,作者还发现在这些人当中,极端近亲繁殖与负面健康后果相关,如肺部功能、视敏度或认知功能下降,这证实了之前的研究发现。此外,作者表明近亲繁殖的后代整体具有更高的疾病风险。
作者指出应该谨慎解读数据,因为样本量较小,而且英国生物样本库可能存在招募偏差(按比例计算,英国生物样本库收录的对象倾向于比其他人口更加健康,具有更高的教育水平)。
摘要:In most human societies, there are taboos and laws banning mating between first- and second-degree relatives, but actual prevalence and effects on health and fitness are poorly quantified. Here, we leverage a large observational study of ~450,000 participants of European ancestry from the UK Biobank (UKB) to quantify extreme inbreeding (EI) and its consequences. We use genotyped SNPs to detect large runs of homozygosity (ROH) and call EI when >10% of an individual’s genome comprise ROHs. We estimate a prevalence of EI of ~0.03%, i.e., ~1/3652. EI cases have phenotypic means between 0.3 and 0.7 standard deviation below the population mean for 7 traits, including stature and cognitive ability, consistent with inbreeding depression estimated from individuals with low levels of inbreeding. Our study provides DNA-based quantification of the prevalence of EI in a European ancestry sample from the UK and measures its effects on health and fitness traits.
(来源:科学网)
特别声明:本文转载仅仅是出于传播信息的需要,并不意味着代表本网站观点或证实其内容的真实性;如其他媒体、网站或个人从本网站转载使用,须保留本网站注明的“来源”,并自负版权等法律责任;作者如果不希望被转载或者联系转载稿费等事宜,请与我们接洽。