|
|
| 突变转座子在真核生物多样性中的进化 | Mobile DNA |
|
论文标题:Evolution of Mutator transposable elements across eukaryotic diversity
期刊:Mobile DNA
作者:Mathilde Dupeyron, Kumar S. Singh, Chris Bass and Alexander Hayward
发表时间:2019/03/21
数字识别码:10.1186/s13100-019-0153-8
原文链接:https://mobilednajournal.biomedcentral.com/articles/10.1186/s13100-019-0153-8?&utm_source=other&utm_medium=other&utm_content=null&utm_campaign=BSCN_2_xw_GeneticsApr_scinet
微信链接:https://mp.weixin.qq.com/s/o8oldDhgrGWXuwngsqBjLw
类突变元件(Mutator-like elements, MULEs)是一类重要的DNA转座子超家族,因为它们拥有以下特征:(i)转座活性大,易于插入或接近基因序列;(i i)高突变能力;(i i i)具有获得宿主基因片段的倾向。因此,MULEs是重要的遗传学研究工具,也是研究宿主转座相互作用的关键系统。然而,以往的许多研究主要关注MULEs对作物和真菌基因组的影响,却缺乏对它们进化机制的探索。来自英国埃克塞特大学的Mathilde Dupeyron研究团队发表在Mobile DNA的文章为您揭开了MULEs进化机制的神秘面纱。
鉴于最近基因组测序数据的大量积累,Mathilde Dupeyron等人重新审视了MULEs的进化问题。为了挖掘MULEs超家族的新成员,他们进行了生物信息学分析并运用系统发育生物学的方法对该超家族的进化普系进行了重建,以更新和进一步研究这一重要DNA 突变体转座因子(Mutator transposable elements , TE)的进化历史。
他们从在线数据库中挖掘MULEs的信息,并将搜索结果与先前已发表研究中检索到的可用转座酶序列相结合,分析结果揭示了两个全新的只存在于节肢动物的MULEs进化枝,这大大地扩展了该组MULEs的组分,为揭开节肢动物基因组的奥秘奠定了基础。
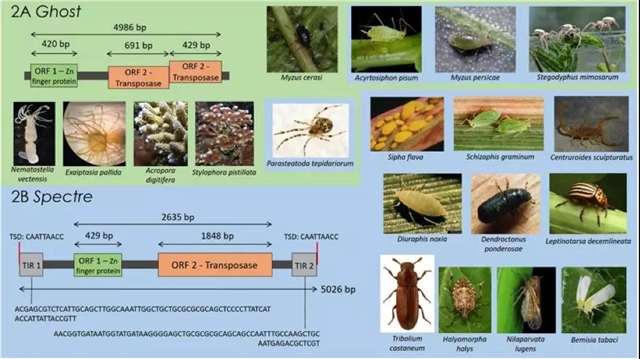
新识别的Ghost和Spectre元件结构示意图,另外还标注了它们各自的宿主范围。
2A:Ghost结构;2B: Spectre结构
植物寄主(主要存在于作物和其他栽培种中)通常被认为是MULEs的主要寄主群体,然而值得注意的是,除了植物寄主,MULEs的存在可能比想象的更为广泛。不过,植物中的MULEs与来自其他寄主群体的MULEs似乎是独在存在的。根据作者先前的研究,他们在6个不同的真核生物群中鉴定出了MULES元件:动物、真菌、变形虫、不等鞭毛类、Parabasalids(一种鞭毛原生生物)和植物,如此广泛的远缘宿主多样性,表明水平转移事件可能在该群体的进化中发挥了重要作用。

一幅描绘MULEs系统发育的图谱。展示了不同发育系统的寄主群体之间MULES的交替联系,除了更多衍生的进化枝,该图显示:这些MULEs元件几乎完全源自植物宿主基因组。
鉴别MULEs的一个有趣的特征便是其异常的末端反向重复序列(Terminal Inverted Repeats,TIR),其长度从14-500 bp不等。研究人员还发现,MULEs的TIR结构在系统发育中的变化很大,并且这种变异似乎与系统发育无关,这种长度差异在组内与组间同样存在。除此之外,他们还根据最新的三代基因组测序结果和新发现的MULEs分支重新对MULEs家族进行了系统学的纠错和分类。总的来讲,Mathilde Dupeyron等人的研究代表了MULEs多样性,宿主范围和进化等研究层面领域的重大进展,并为其他待发现的MULEs元件的分类提供了理论框架。
摘要:
Background
Mutator-like elements (MULEs) are a significant superfamily of DNA transposons on account of their: (i) great transpositional activity and propensity for insertion in or near gene sequences, (ii) their consequent high mutagenic capacity, and, (iii) their tendency to acquire host gene fragments. Consequently, MULEs are important genetic tools and represent a key study system for research into host-transposon interactions. Yet, while several studies have focused on the impacts of MULEs on crop and fungus genomes, their evolution remains poorly explored.
Results
We perform comprehensive bioinformatic and phylogenetic analyses to address currently available MULE diversity and reconstruct evolution for the group. For this, we mine MULEs from online databases, and combine search results with available transposase sequences retrieved from previously published studies. Our analyses uncover two entirely new MULE clades that contain elements almost entirely restricted to arthropod hosts, considerably expanding the set of MULEs known from this group, suggesting that many additional MULEs may await discovery from further arthropod genomes. In several cases, close relationships occur between MULEs recovered from distantly related host organisms, suggesting that horizontal transfer events may have played an important role in the evolution of the group. However, it is apparent that MULEs from plants remain separate from MULEs identified from other host groups. MULE structure varies considerably across phylogeny, and TIR length is shown to vary greatly both within and between MULE groups. Our phylogeny suggests that MULE diversity is clustered in well-supported groups, typically according to host taxonomy. With reference to this, we make suggestions on how MULE diversity can be partitioned to provide a robust taxonomic framework.
Conclusions
Our study represents a considerable advance in the understanding of MULE diversity, host range and evolution, and provides a taxonomic framework for the classification of further MULE elements that await discovery. Our findings also raise a number of questions relating to MULE biology, suggesting that this group will provide a rich avenue for future study.
阅读论文全文请访问:
https://mobilednajournal.biomedcentral.com/articles/10.1186/s13100-019-0153-8?&utm_source=other&utm_medium=other&utm_content=null&utm_campaign=BSCN_2_xw_GeneticsApr_scinet
期刊介绍:
Mobile DNA (https://mobilednajournal.biomedcentral.com/, 5.891 -2-year Impact Factor, 3.957 -5-year Impact Factor)is an open access, peer-reviewed journal that publishes articles providing novel insights into DNA rearrangements in all organisms, ranging from transposition and other types of recombination mechanisms to patterns and processes of mobile element and host genome evolution. In addition, the journal will consider articles on the utility of mobile genetic elements in biotechnological methods and protocols.
(来源:科学网)
特别声明:本文转载仅仅是出于传播信息的需要,并不意味着代表本网站观点或证实其内容的真实性;如其他媒体、网站或个人从本网站转载使用,须保留本网站注明的“来源”,并自负版权等法律责任;作者如果不希望被转载或者联系转载稿费等事宜,请与我们接洽。